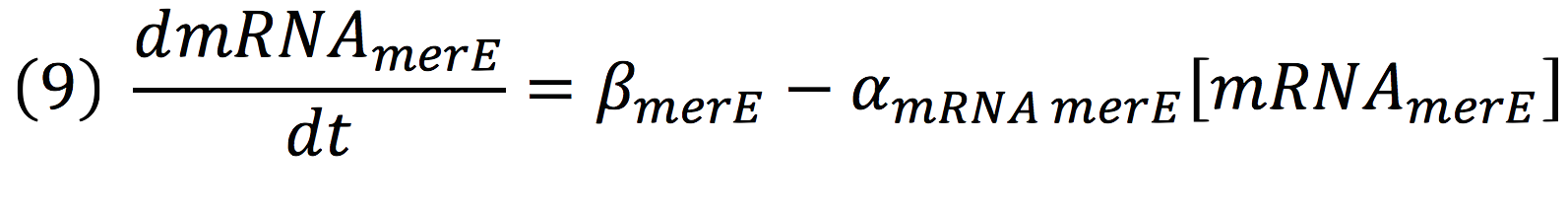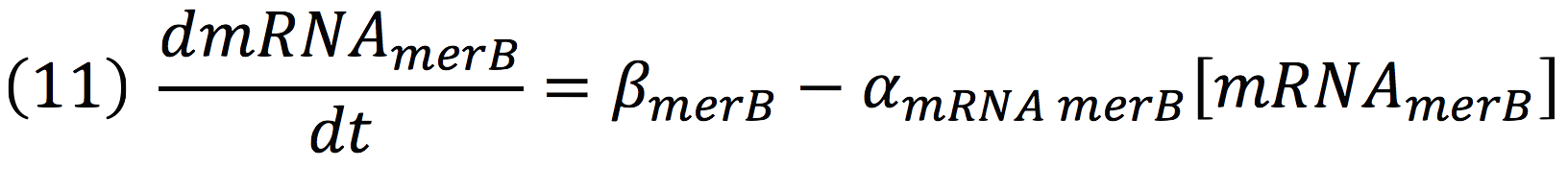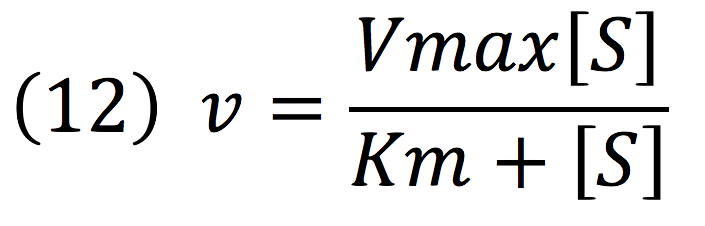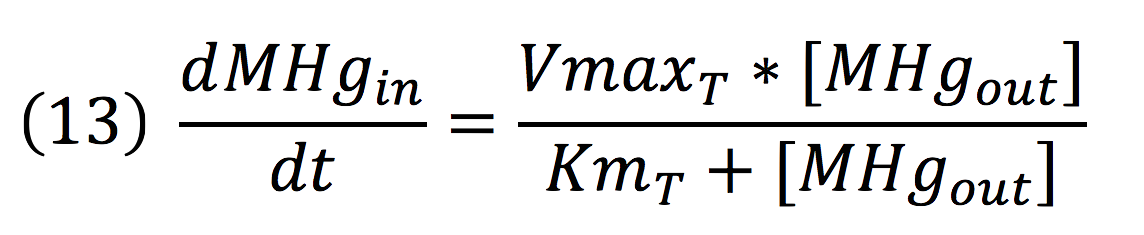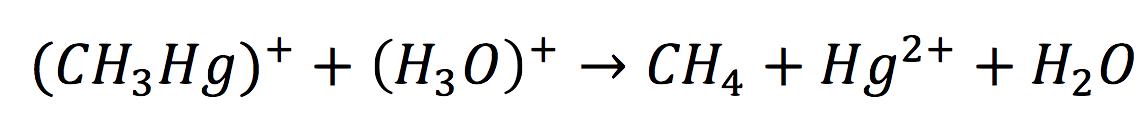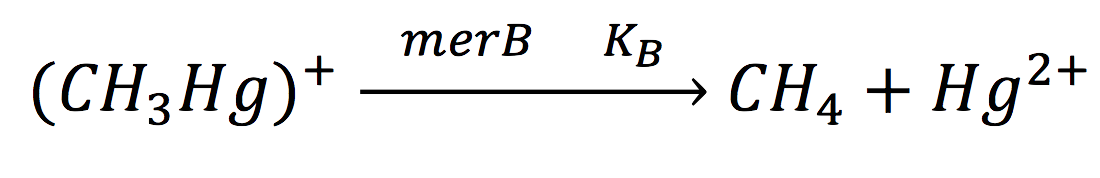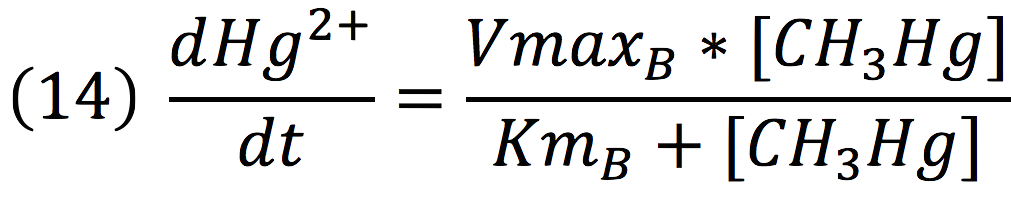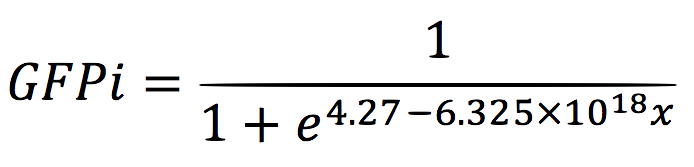Team:BIOSINT Mexico/Bioaccumulation
Mercury Bioaccumulation
Description
Detection
Heavy metal-mediated toxicity has always been one of the greatest obstacles against survival of microorganisms. Bacteria, however, have evolved a great spectrum of mechanisms to deal with such impediment. The genetic system “mer operon” is the only bacterial metal resistance system with high yield transformation of its toxic target into volatile non-toxic forms. Basically, protein products of mer genes efficiently utilize the high affinity of Hg2+ towards cysteine residues in proteins for their enzymatic degradation and transportation (Mathema VB et al, 2011). As there are various genes involved in mer operon that constitute the different parts of the mercurial environment detoxification mechanism, we selected from these pull of genes those that could be better suited for our chassis.
Transportation mechanism
The mechanism of transport of Hg2+ and CH3Hg inherently across the bacterial membrane is mediated by MerC, MerE, MerT and MerP. Even though among the four transporters, MerC showed highest potential for Hg2+ transport across the bacterial membrane (Sone Y et al, 2013), for broader purposes of this work, MerE will was picked to evaluate its effect on bioaccumulation of methylmercury in A. thaliana.
In addition MerE is able to transport methylmercury and ions of mercury ions across bacterial cytoplasmic membranes. When arabidopsis thaliana works with the gene MerE it has been demonstrated to gain resistance to methylmercury and to the mercury ions, promoting the transport and accumulation of this two elements and eventually facilitating methyl mercury phytoremediation (Sone Y. et al, 2013).
Reduction
MerB gene codes for organomercurial lyase, that catalyzes the protonolysis of the carbon-mercury bond.(Bizily S.P et al, 1999) Thus the products of this reaction are a less toxic inorganic Hg2+ species and a reduced carbon compound.
In presence of MerB: R-CH2-Hg++H+ R-CH3+Hg2+
“The kinetics of the MerB-catalyzed reaction could be constrained by the rate of diffusion of organomercurial substrates from cellular membrane systems to sites of catalysis or by the rate of diffusion of the product, Hg2+, away from the enzyme” (Bizily S.P et al, 1999).
MerB gives resistance to organomercurial lyase allowing plants to efficiently protonolyze organic mercury producing a more tolerable mercury species (Bizily S.P et al, 1999) .
Hg2+ is generated by MerB within the citoplasm in the presence of cellular proteins, it remains paradoxical that plants with MerE tolerate more organic mercury than wild-type plants (Bizily S.P et al, 1999). The reason of this situation is that the lipid solubility of organic mercury gives access to mitochondria and chloroplasts, where it may affect essential oxidative and photosynthetic electron transport chains. It is also probable that cytoplasmic chelators dins and sequester Hg2+ (Bizily S.P et al, 1999) in preference of organomercurials. Sustaining, in this case, the function of our module by naturally selecting methyl mercury over mercury´s ionic form and inducing our system more continously.
Modeling
Equations for mer Operon production
For the mer operon expression we start from the reaction
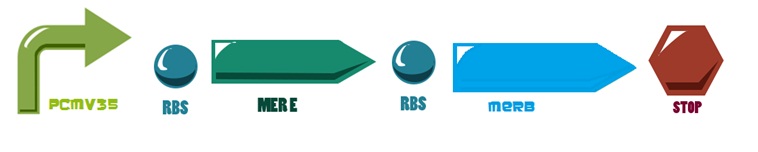
Of all the genes of this operon we used two specific proteins. merE is a transport protein in charge of the active transport od methyl-mercury from the environment of the cell to its interior. Once in the inner cytoplasm, merB protein attacks the molecule and breaks the covalent bond releasing methane and Hg2+(s) to the cell. Methane is metabolized and the quicksilver is accumulated.
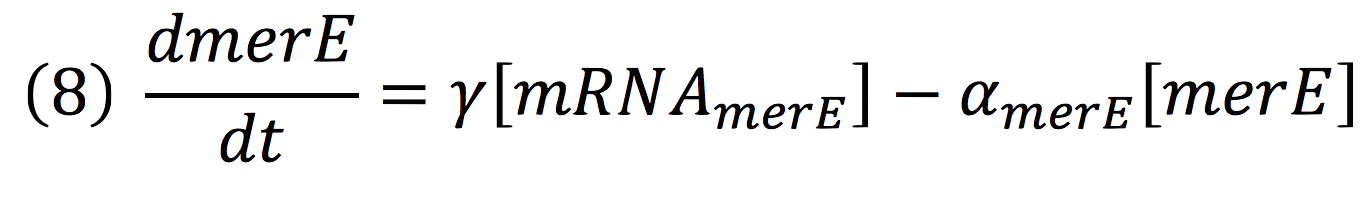
Then, for the merE production rate we know that
And, as mer operon is attached to a minimal constitutive promoter (PCMV), the production rate of the messenger molecule is
The same process is applied to merB from the operon. Therefore:
Equations for Hg transportation
Methyl mercury transportation is mediated by the merE enzyme from the mer operon. The metal present in the environment of the cell is transported to the inner plant cell following the reaction:
Where the metallic molecule is taken from the outside (MHgout) to the inside (MHgin) by a constant of reaction of KT and where K-T equals zero because the mercury inside cannot return to the exterior of the cell by the same reaction, given that mercury transportation mediated by merE is a one way process.
Since the reaction is mediated by an enzyme, it could be expressed by:
Then, it is possible to model the process just as any other enzyme mediated reaction. For this purpose, we use the Michaelis-Menten equation.
Where v is the reaction rate, Vmax is the maximum rate achieved by the reaction at saturating substrate concentrations, S is the substrate and Km is the Michaelis constant for the reaction, or the substrate concentration when the system achieves half of Vmax. Therefore, adapting this for our process, we have:
Equations for mercury bioremediation
Once in the interior of the cell, under slightly acid conditions, methyl mercury decays on methane and metallic mercury, by the following reaction:
Since the rate of the reaction in normal conditions is very low, we use merB to accelerate this rate. Therefore, the reaction can be described as:
Where methyl mercury is lysed by the merB protein at a rate that goes by the Michaelis-Menten equation:
Where the methyl mercury concentration is the concentration of the molecule inside the cell, described in the last step.
Results
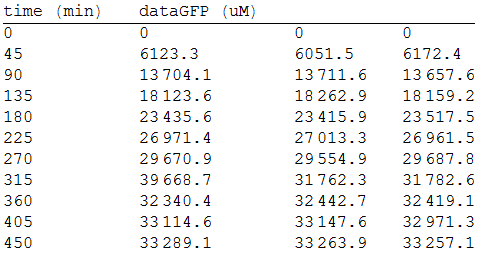
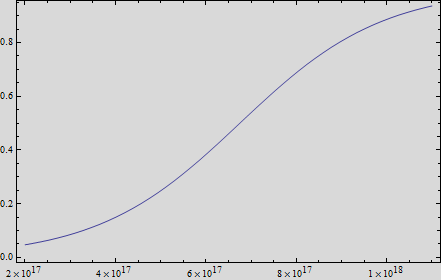
In order to measure the mer operon expression fused the composite to a GFP reporter and analized the bioremediation by measuring the change of the concentration of methyl mercury in the media. First, we measured the GFP fluorescense intensity every 45 minutes.
As we make three repetitions, we take the mean values as input data for the regression. The analized data was:
For the regression, we used the Wolfram Mathematica software and obtained the equation:
Thus, if we plot the equation we obtain that:
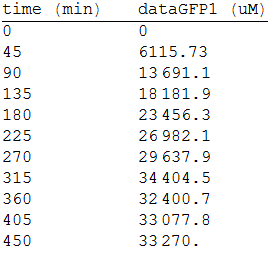
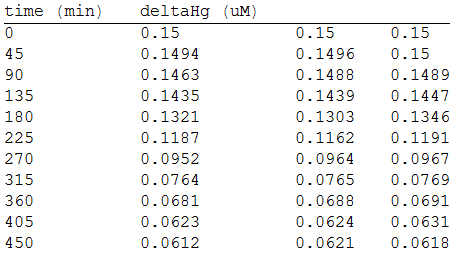
And, as we see in the graph, GFP expresses directly proportional to time, given the strenght of the promoter upstream. Then, as GFP expresses at aproximately the same rate that the mer operon proteins, it is posible to infer that the GFP concentration given any time is the same as the mer operon proteins concentration attached upstream. The change in the concentration of methyl mercury was also measured. We choose to measure it instead of the mercury accumulated inside the cells because it was easier. The collected data is provided in the table:
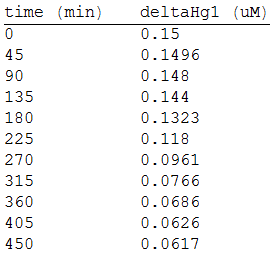
As in the other experiments, we analyzed the mean values of the concentration in order to make the regression:
Obtaining the polynomial equation:
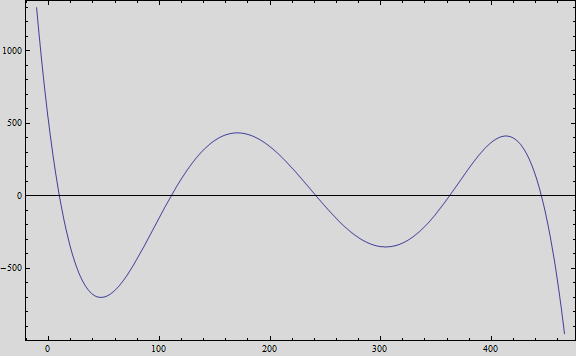
And, if we plot the equation we obtain the following graph:
That, at short term, fits the behavior of the expected values.
References
[1]Bizily S., et al. (1999). Phytoremediation of methylmercury pollution: MerB expression in Arabidopsis Thaliana confers resistance to organomercurials. Proc. Natl. Acad. Vol. 96, pp. 6806-6813, June1999.
[2]Sone Y., et al. (2010). Roles played by MerE and MerT in the transport of inorganic and organic mercury compounds in Gram-negative Bacteria. Journal of Health Science, 56(1) 123-127 2010.
[3]Sone Y., et al. (2013). Increase methylmercury accumulation in Arabidopsis thaliana expressing bacterial broad-spectrum mercury transporter MerE. AMB Express 2013 3:52.
[4] Barkay, T., Miller, S. and Summers, A. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS microbiology reviews, 27(2-3), pp.355--384.
 "
"