Team:Aberdeen Scotland/Parts/Device
From 2014.igem.org
| (5 intermediate revisions not shown) | |||
| Line 53: | Line 53: | ||
<li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2001">Bba_K1352001</a></li> | <li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2001">Bba_K1352001</a></li> | ||
<li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2002">Bba_K1352002</a></li> | <li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2002">Bba_K1352002</a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2003">Bba_K1352003</a></li> | ||
<li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2004">Bba_K1352004</a></li> | <li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2004">Bba_K1352004</a></li> | ||
<li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2006">Bba_K1352006</a></li> | <li><a href="https://2014.igem.org/Team:Aberdeen_Scotland/Parts/_2006">Bba_K1352006</a></li> | ||
| Line 70: | Line 71: | ||
<!-- SECTION HEAD --> | <!-- SECTION HEAD --> | ||
<div class="t_overview"> | <div class="t_overview"> | ||
| - | <h1> | + | <h1>Detector Test Data</h1> |
<!-- <h3></h3> --> | <!-- <h3></h3> --> | ||
</div> <br class="clear"> <!-- END OF HEAD --> | </div> <br class="clear"> <!-- END OF HEAD --> | ||
| Line 76: | Line 77: | ||
<!-- PAGE CONTENT --> | <!-- PAGE CONTENT --> | ||
<div class="main_content"> | <div class="main_content"> | ||
| - | <p> | + | <p>The initial concept of the fluorescence detector device had a very rough design. Thus it was built from bits and pieces in order to do a preliminary test and |
| - | + | verify its viability.</p> | |
| - | + | ||
| - | + | <img src="https://static.igem.org/mediawiki/2014/a/a0/Rough_set.jpg"> | |
| - | + | ||
| - | <img src="https://static.igem.org/mediawiki/2014/ | + | <p>The preliminary data from the tests showed that the detector was able to differentiate between GFP producing and non-producing bacteria.</p> |
| - | + | ||
| + | <img src="https://static.igem.org/mediawiki/2014/e/e9/Lb_vs_gfp.png"> | ||
| + | |||
| + | <p>Fig.1 Series dilutions to estimate detector sensitivity</p> | ||
| + | |||
| + | <p>From Fig.1 we can see that the detector can easily distinguish the GFP producing culture from below 1/10 dilutions. Thus we continued on and improved the design by soldering some of the circuit and making it more compact. Then we made more test with the Sender and Receiver bacteria.</p> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/3/33/Send_rec_vs.png"> | ||
| + | <p>Fig.2 Detector response to Sender and Receiver cultures</p> | ||
| + | |||
| + | <p>Again the detector is sensitive enough to pick up the GFP in the Receiver.</p> | ||
| + | |||
| + | <p>For the final test, we made clean overnight cultures of the Receiver and Sender and mixed them in a growing culture. This was done to see after how long the detector is going to pick up the GFP production. Hence, if our diagnostic method is going to work.</p> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/7/79/Resp_time.png"> | ||
| + | <p>Fig.3 Detector response to QS GFP-producing Receiver and Sender culture, normalised against LB medium</p> | ||
| + | |||
| + | <p><b>Finally, the simple detector proved capable of detecting the QS culture, thus confirming our design success.</b></p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> <br class="clear"> <!-- END OF PAGE CONTENT --> | </div> <br class="clear"> <!-- END OF PAGE CONTENT --> | ||
Latest revision as of 02:31, 18 October 2014
Detector Test Data
The initial concept of the fluorescence detector device had a very rough design. Thus it was built from bits and pieces in order to do a preliminary test and verify its viability.
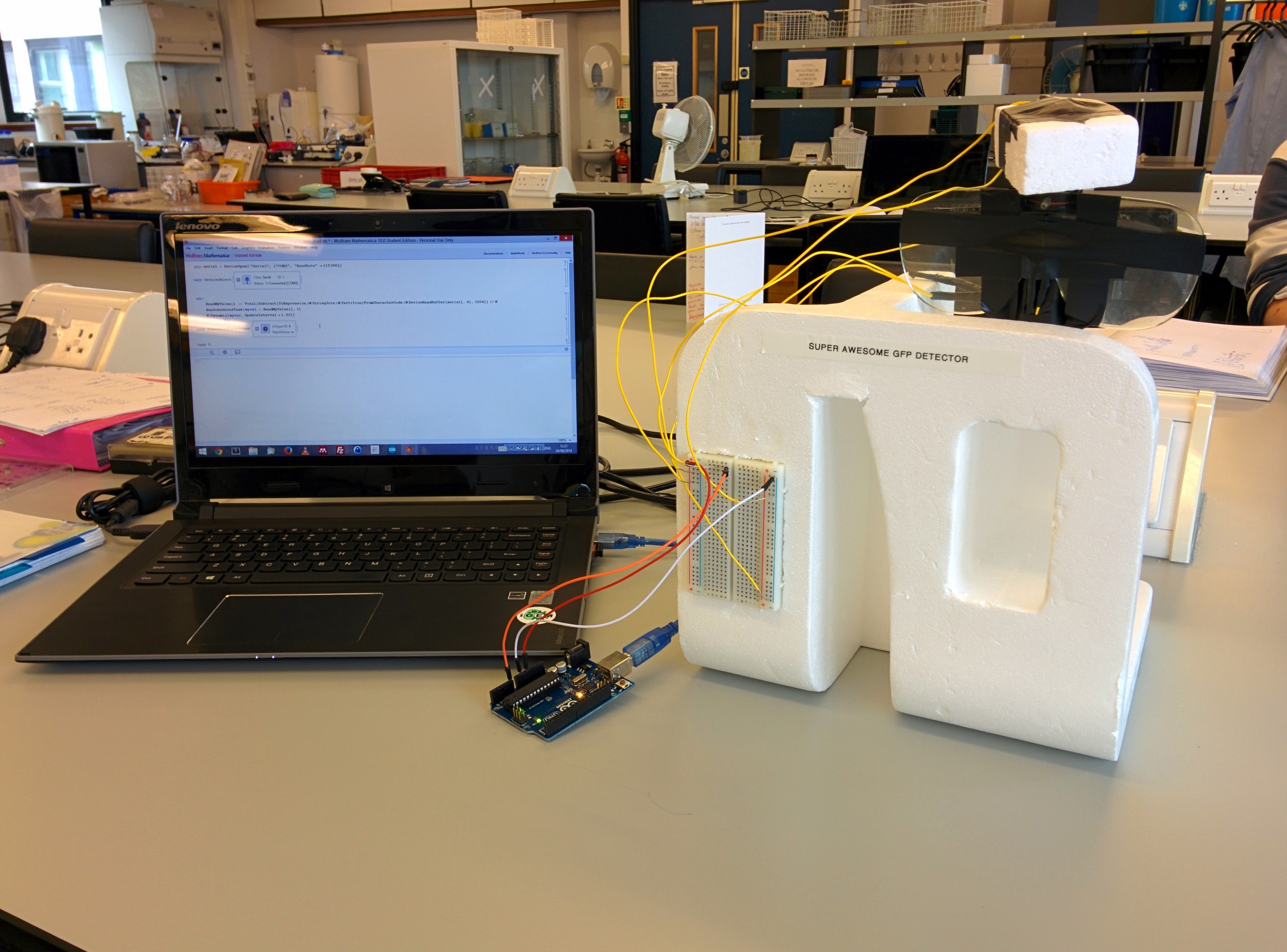
The preliminary data from the tests showed that the detector was able to differentiate between GFP producing and non-producing bacteria.

Fig.1 Series dilutions to estimate detector sensitivity
From Fig.1 we can see that the detector can easily distinguish the GFP producing culture from below 1/10 dilutions. Thus we continued on and improved the design by soldering some of the circuit and making it more compact. Then we made more test with the Sender and Receiver bacteria.

Fig.2 Detector response to Sender and Receiver cultures
Again the detector is sensitive enough to pick up the GFP in the Receiver.
For the final test, we made clean overnight cultures of the Receiver and Sender and mixed them in a growing culture. This was done to see after how long the detector is going to pick up the GFP production. Hence, if our diagnostic method is going to work.

Fig.3 Detector response to QS GFP-producing Receiver and Sender culture, normalised against LB medium
Finally, the simple detector proved capable of detecting the QS culture, thus confirming our design success.
 "
"






