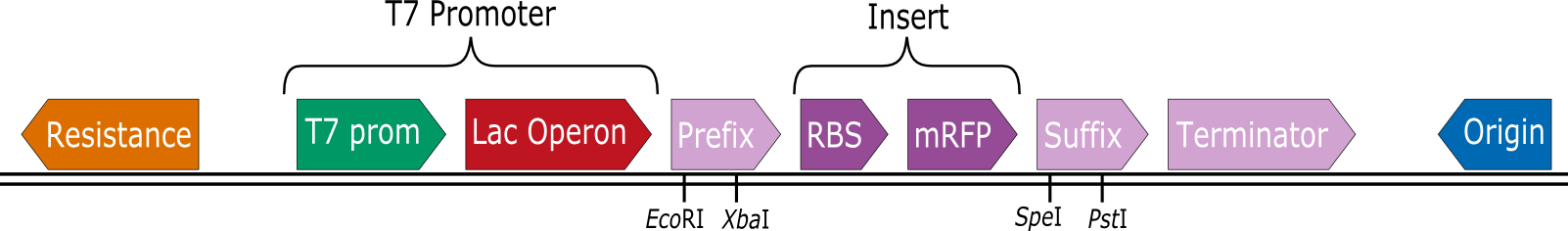
Team:Aachen/Collaborations/Heidelberg
From 2014.igem.org
(Difference between revisions)
m (→The Team Heidelberg expression vectors) |
|||
| Line 43: | Line 43: | ||
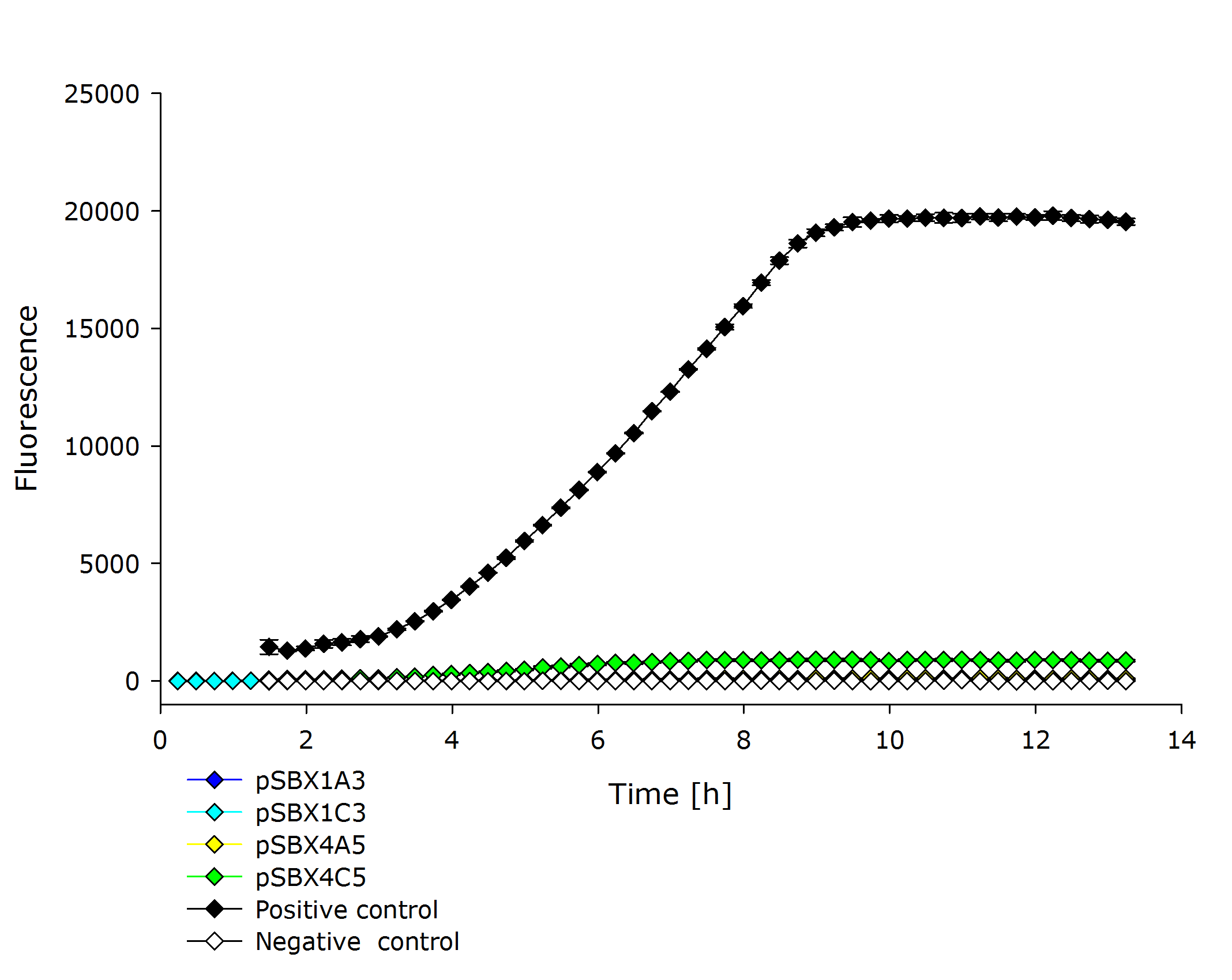
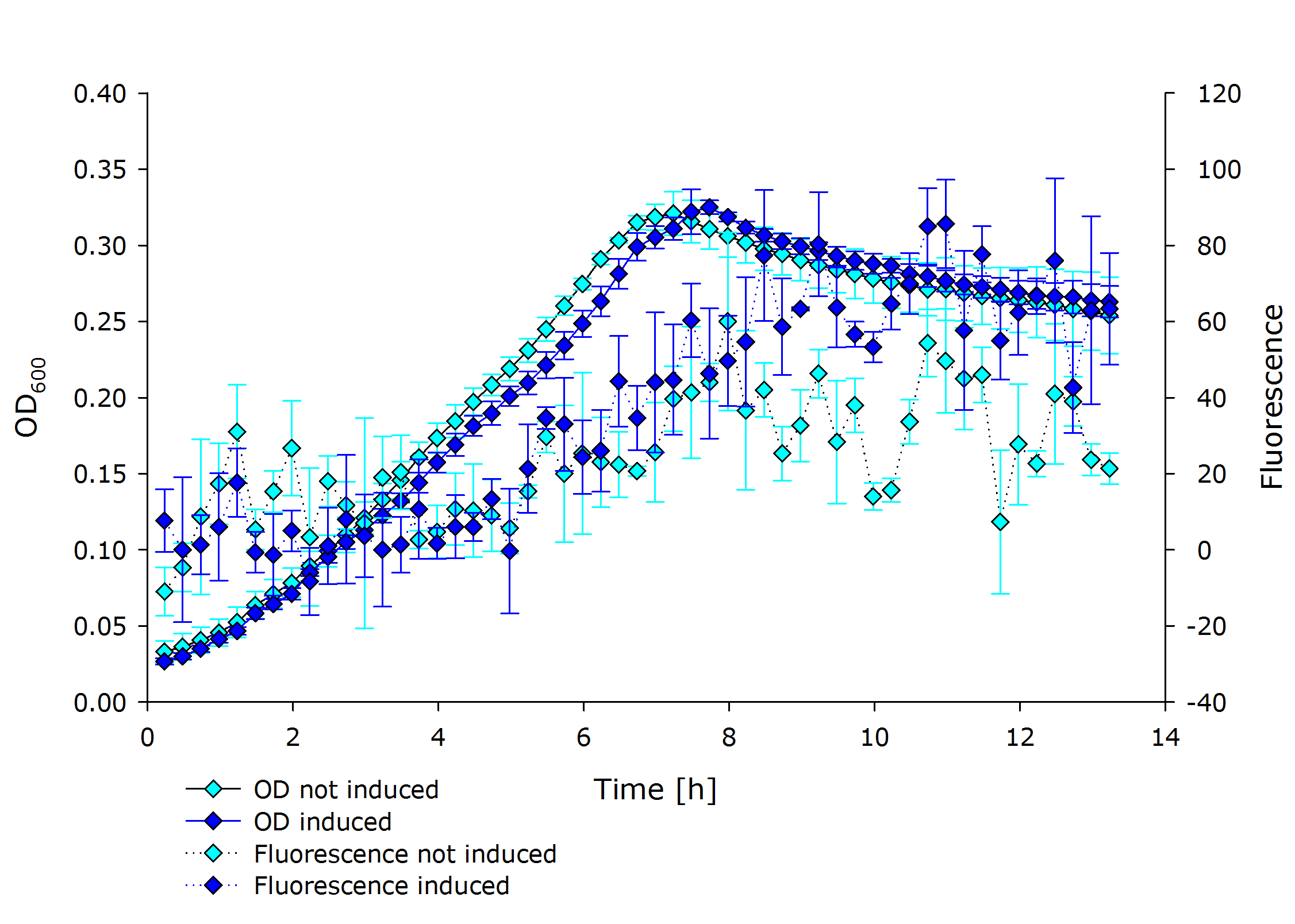
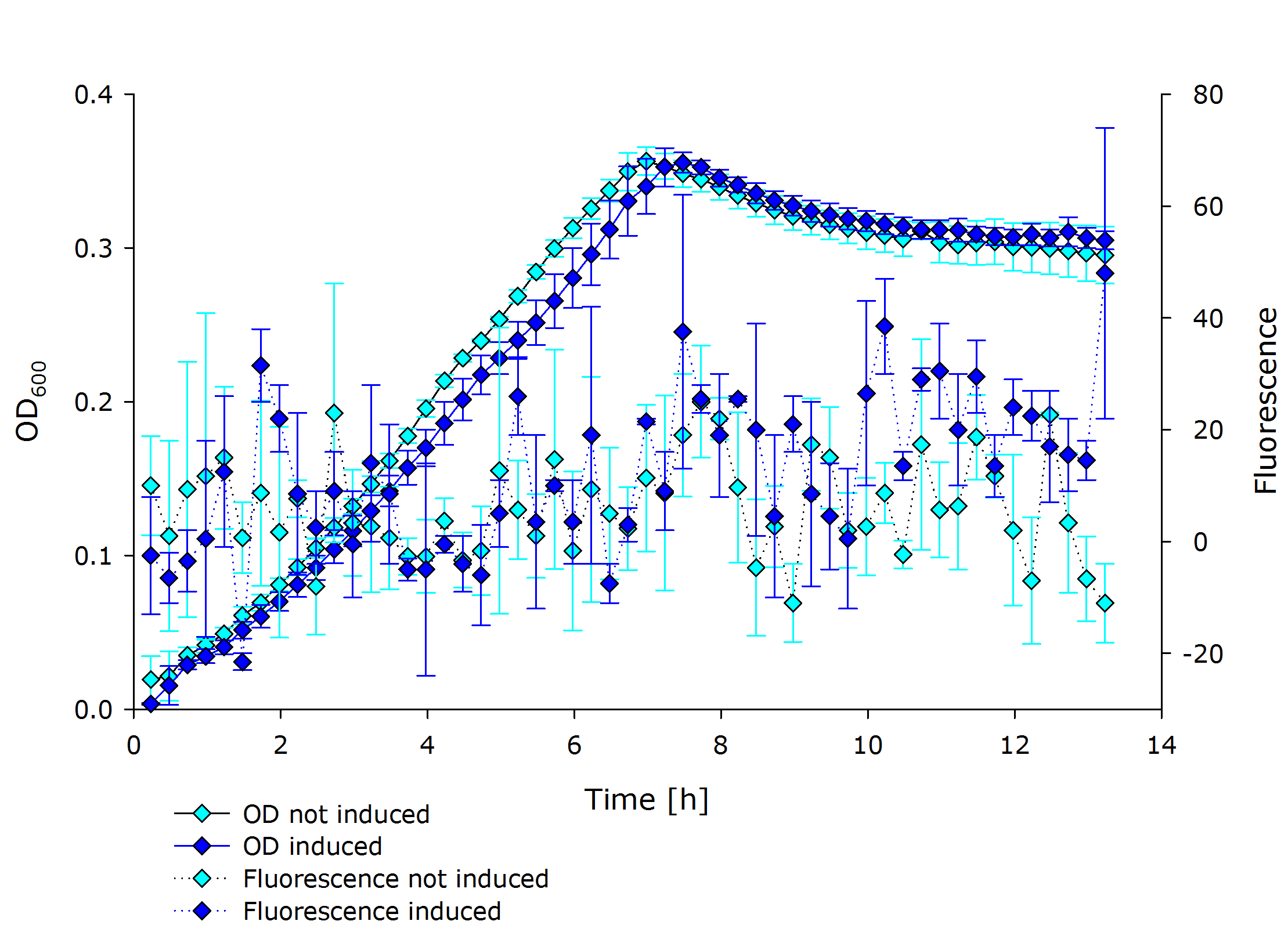
{{Team:Aachen/Figure|Aachen 17-10-14 PSBX1C3 iFG.PNG|||width=200px}} | {{Team:Aachen/Figure|Aachen 17-10-14 PSBX1C3 iFG.PNG|||width=200px}} | ||
{{Team:Aachen/Figure|Aachen 17-10-14 PSBX4A5 iFG.PNG|||width=200px}} | {{Team:Aachen/Figure|Aachen 17-10-14 PSBX4A5 iFG.PNG|||width=200px}} | ||
| - | {{Team:Aachen/Figure|Aachen 17-10-14 PSBX4C5 iFG.PNG|||width=200px}} | + | {{Team:Aachen/Figure|Aachen 17-10-14 PSBX4C5 iFG.PNG|||width=200px}}, |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | blabla | ||
| + | |||
| + | |||
| + | |||
| + | <div class="figure" style="float:center; margin: 10px 10px 10px 40px; border:0px solid #aaa;width:600px;padding:0px;"> | ||
| + | {| | ||
| + | |{{Team:Aachen/Test/GalleryHeader}} | ||
| + | <html> | ||
| + | <div id="slider1_container" style="position: relative; width: 600px; height: 500px; background-color: #000; overflow: hidden; "> | ||
| + | |||
| + | |||
| + | <div u="loading" style="position: absolute; top: 0px; left: 0px;"> | ||
| + | <div style="filter: alpha(opacity=70); opacity:0.7; position: absolute; display: block; | ||
| + | background-color: #000; top: 0px; left: 0px;width: 100%;height:100%;"> | ||
| + | </div> | ||
| + | <div style="position: absolute; display: block; background: url(../img/loading.gif) no-repeat center center; | ||
| + | top: 0px; left: 0px;width: 100%;height:100%;"> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <!-- Slides Container --> | ||
| + | <div u="slides" style="cursor: move; position: absolute; left: 0px; top: 0px; width: 600px; height: 500px; | ||
| + | overflow: hidden;"> | ||
| + | <div> | ||
| + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/thumb/a/a6/Aachen_17-10-14_Summary_iFG.PNG/754px-Aachen_17-10-14_Summary_iFG.PNG"/></a> | ||
| + | </div> | ||
| + | <div> | ||
| + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/thumb/0/04/Aachen_17-10-14_PSBX1A3_iFG.PNG/800px-Aachen_17-10-14_PSBX1A3_iFG.PNG"/></a> | ||
| + | </div> | ||
| + | <div> | ||
| + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/thumb/8/8c/Aachen_17-10-14_PSBX1C3_iFG.PNG/800px-Aachen_17-10-14_PSBX1C3_iFG.PNG"/></a> | ||
| + | </div> | ||
| + | <div> | ||
| + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/thumb/0/08/Aachen_17-10-14_PSBX4A5_iFG.PNG/800px-Aachen_17-10-14_PSBX4A5_iFG.PNG"/></a> | ||
| + | </div> | ||
| + | <div> | ||
| + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/thumb/9/98/Aachen_17-10-14_PSBX4C5_iFG.PNG/796px-Aachen_17-10-14_PSBX4C5_iFG.PNG"/></a> | ||
| + | </div> | ||
| + | </div> | ||
| + | <a style="display: none" href="http://www.jssor.com">jquery slider example</a> | ||
| + | <div u="navigator" class="jssorb13" style="position: absolute; bottom: 16px; right: 6px;"> | ||
| + | <div u="prototype" style="POSITION: absolute; WIDTH: 21px; HEIGHT: 21px;"></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <link rel="stylesheet" href="https://2014.igem.org/Team:Aachen/Scripts/highlight.pack.default.css?action=raw&ctype=text/css"> | ||
| + | <script src="https://2014.igem.org/Team:Aachen/Scripts/highlight.pack.js?action=raw&ctype=text/javascript"></script> | ||
| + | <script>hljs.initHighlightingOnLoad();</script> | ||
| + | </body> | ||
| + | </html> | ||
| + | |- | ||
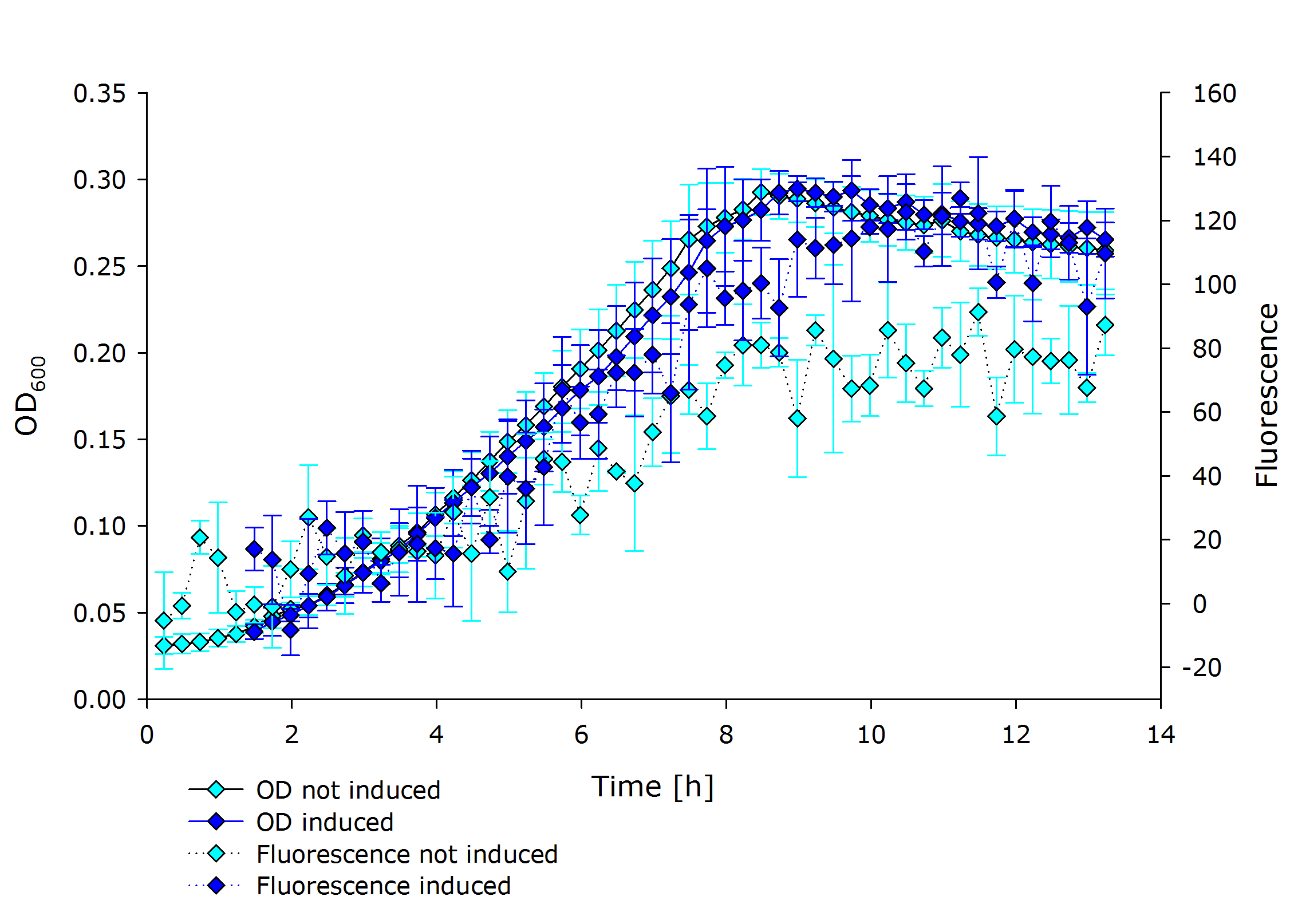
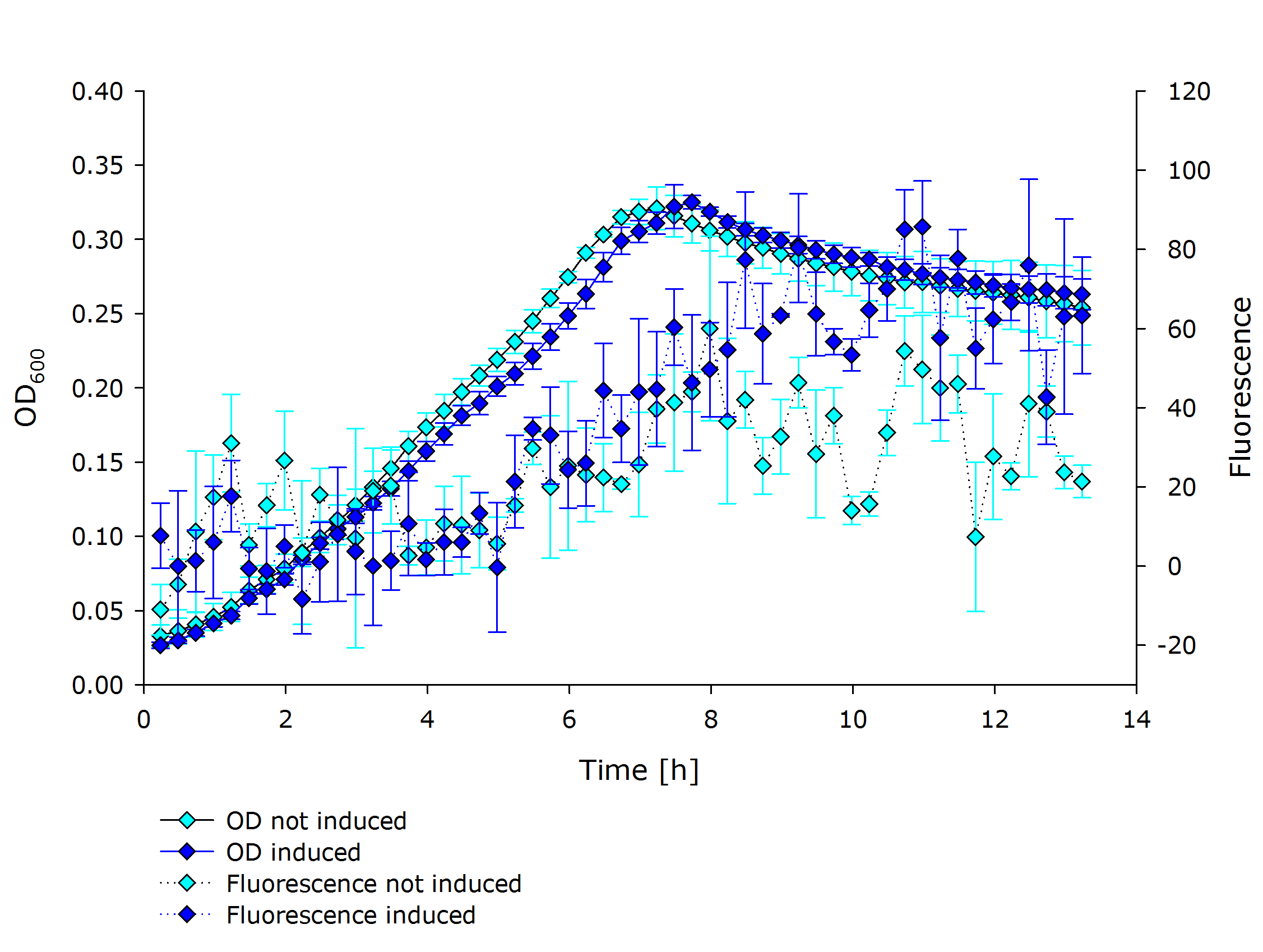
| + | |'''Fluorescence data of pSBX cultivation'''<br />todo.. | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | blabla | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
{{Team:Aachen/Footer}} | {{Team:Aachen/Footer}} | ||
Revision as of 20:03, 17 October 2014
|
 "
"