|
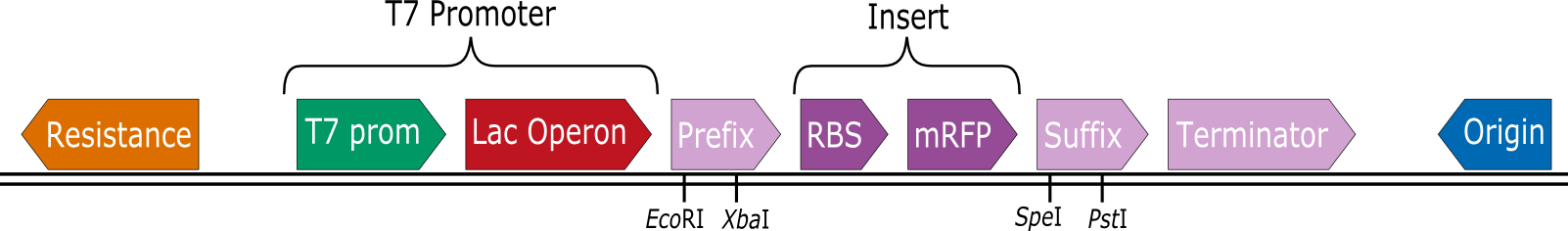
The iGEM team Heidelberg constructed a set of expression vectors that contain a T7 promoter prior to the BioBrick prefix. This way RBS-CDS parts can be cloned into the vectors by standard BioBrick assembly and then expressed in BL21(DE3) by induction with IPTG.
In collaboration with the iGEM Team Heidelberg the iGEM Team Aachen tested 8 of these vectors for their expression capabilities.
We successfully cloned and expressed one of our fusion proteins K139020 in a pSBX1A3 vector. You can read more about the background of this expression on the Galectin-3 page.
Attempting to Characterize the pSBX vectors
These are the tested constructs:
| Plasmid | Backbone | Insert | Length vector [bp] | Length Insert [bp] | Resistance
|
| pSBX1A3-BBa_13507 | pSBX1A3 (high copy) | BBa_13507 | 3067 | 1226 | 50 mg/ml Ampicilin
|
| pSBX1C3-BBa_13507 | pSBX1C3 (high copy) | BBa_13507 | 2982 | 1226 | 35 mg/ml Chloramphenicol
|
| pSBX1K3-BBa_13507 | pSBX1K3 (high copy) | BBa_13507 | 3116 | 1226 | 50 mg/ml Kanamycin
|
| pSBX1T3-BBa_13507 | pSBX1T3 (high copy) | BBa_13507 | 3322 | 1175 | 10 mg/ml Tetracyclin
|
| pSBX4A5-BBa_13507 | pSBX4A5 (low copy) | BBa_13507 | 4307 | 1192 | 50 mg/ml Ampicilin
|
| pSBX4C5-BBa_13507 | pSBX4C5 (low copy) | BBa_13507 | 4127 | 1186 | 15 mg/ml Chloramphenicol
|
| pSBX4K5-BBa_13507 | pSBX4K5 (low copy) | BBa_13507 | 4331 | 1192 | 50 mg/ml Kanamycin
|
We also used an mRFP selection marker with a constant expression of mRFP as a positive control.
To find out more about the Results of the characterization check out the the collaboration page of Team Heidelberg.
|
 "
"