Team:Austin Texas/kit
From 2014.igem.org
| Line 75: | Line 75: | ||
__TOC__ | __TOC__ | ||
| - | + | <h1>Kit Introduction</h1> | |
In recent years the ability to expand the genetic code has been made possible by re-coding the amber stop codon, UAG, via the use of modified tRNA synthetase/tRNA pairs. These synthetase/tRNA pairs act together to charge the tRNA with a non-canonical amino acid (ncAA), an amino acid that is not one of the 20 amino acids normally encoded by a codon. While the library of ncAA synthetase/tRNA pairs continues to grow, the properties of each pairing have yet to be systematically characterized using a standardized methodology. | In recent years the ability to expand the genetic code has been made possible by re-coding the amber stop codon, UAG, via the use of modified tRNA synthetase/tRNA pairs. These synthetase/tRNA pairs act together to charge the tRNA with a non-canonical amino acid (ncAA), an amino acid that is not one of the 20 amino acids normally encoded by a codon. While the library of ncAA synthetase/tRNA pairs continues to grow, the properties of each pairing have yet to be systematically characterized using a standardized methodology. | ||
| Line 84: | Line 84: | ||
| - | + | <h2>Motivation</h2> | |
[[File:Translation of ncAA 10-16-14.png|300px|thumb|right|<b>Figure 1</b> Translation of non-canonical amino acids.]] | [[File:Translation of ncAA 10-16-14.png|300px|thumb|right|<b>Figure 1</b> Translation of non-canonical amino acids.]] | ||
| Line 96: | Line 96: | ||
| - | + | ||
| + | <h1>Background</h1> | ||
The genetic code is a composition of 20 highly conserved amino acids that are essential to all organisms on Earth. While the genetic code is specific, it is also degenerate, meaning that more than one codon can encode for the incorporation of a specific amino acid. For example, there are six serine codons and three stop codons (called amber, ochre, and opal). By recoding one of the redundant codons, the recoded codon can signal for the incorporation of a non-canonical amino acid (ncAA) rather than the codon's original usage. Of the three stop codons, the amber codon is the least abundant and thus, the easiest and most efficient to recode. | The genetic code is a composition of 20 highly conserved amino acids that are essential to all organisms on Earth. While the genetic code is specific, it is also degenerate, meaning that more than one codon can encode for the incorporation of a specific amino acid. For example, there are six serine codons and three stop codons (called amber, ochre, and opal). By recoding one of the redundant codons, the recoded codon can signal for the incorporation of a non-canonical amino acid (ncAA) rather than the codon's original usage. Of the three stop codons, the amber codon is the least abundant and thus, the easiest and most efficient to recode. | ||
| Line 106: | Line 107: | ||
Complications arise when the genetic code is recoded. In a normal bacterium, release factor RF1 is responsible for terminating translation when the ribosome reaches the amber stop codon. To avoid termination at a UAG amber codon, a strain of ''E. coli'' was engineered by the Church and Isaacs groups using MAGE and CAGE to remove all of the amber codons from the genome and knock out the RF1 gene (Isaacs et al. 2011). The resulting strain, called "amberless" ''E. coli'', has all amber codons free to code for any ncAA. When both are present during translation, a synthetase with mutations that allow for the acceptance of an ncAA, charges that ncAA onto its orthogonal tRNA with the amber codon's anticodon, CUA. | Complications arise when the genetic code is recoded. In a normal bacterium, release factor RF1 is responsible for terminating translation when the ribosome reaches the amber stop codon. To avoid termination at a UAG amber codon, a strain of ''E. coli'' was engineered by the Church and Isaacs groups using MAGE and CAGE to remove all of the amber codons from the genome and knock out the RF1 gene (Isaacs et al. 2011). The resulting strain, called "amberless" ''E. coli'', has all amber codons free to code for any ncAA. When both are present during translation, a synthetase with mutations that allow for the acceptance of an ncAA, charges that ncAA onto its orthogonal tRNA with the amber codon's anticodon, CUA. | ||
| - | + | <h1>Experimental Design and Method</h1> | |
| - | + | <h1>Plasmids</h1> | |
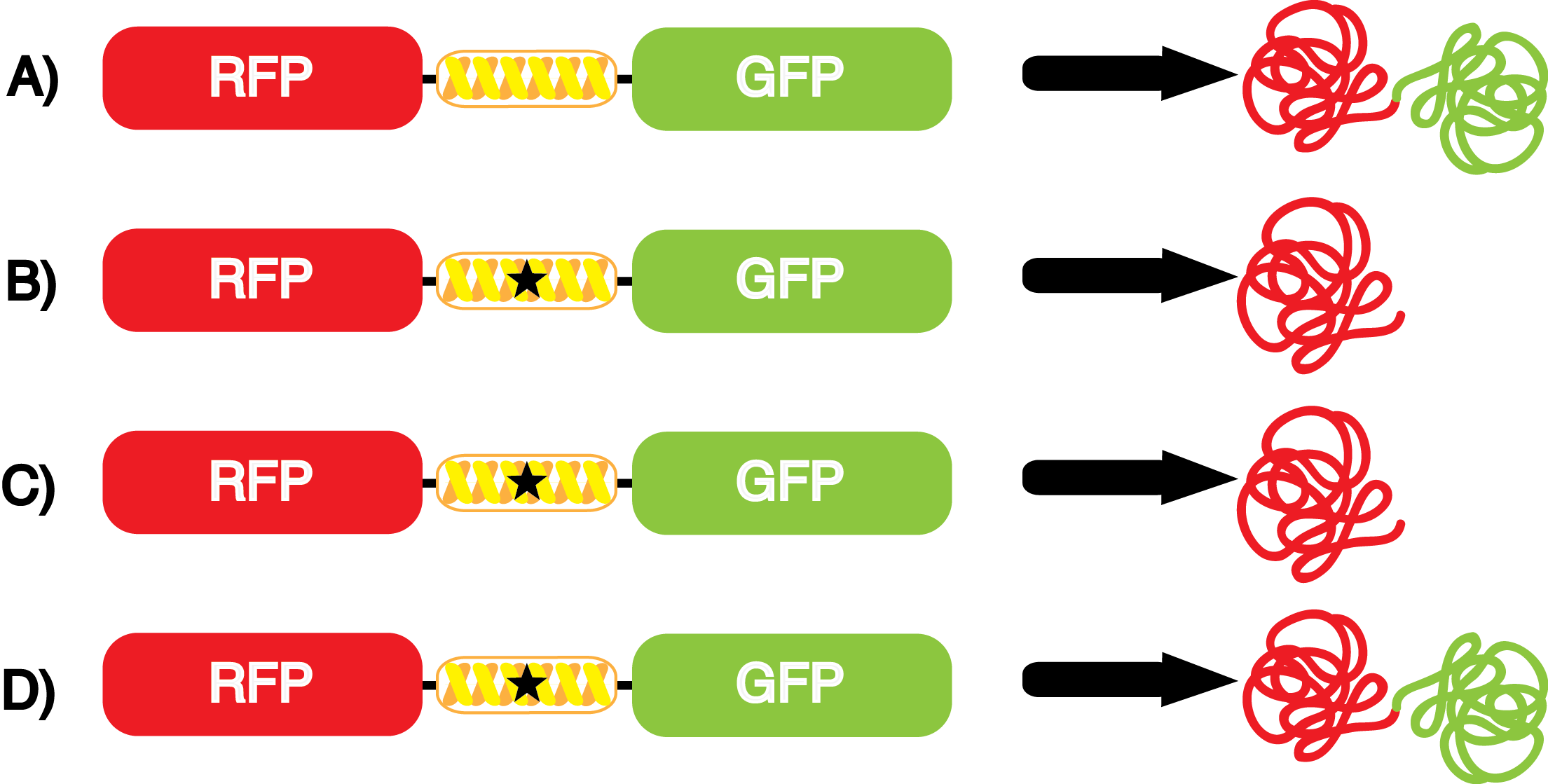
[[File:Kit Plasmid Alignment 10-14-14.png|570px|thumb|right|<b>Figure 2</b> | [[File:Kit Plasmid Alignment 10-14-14.png|570px|thumb|right|<b>Figure 2</b> | ||
| Line 122: | Line 123: | ||
| - | + | ||
| + | |||
| + | <h2>Using The Kit</h2> | ||
To use the kit properly, each culture must have two of the three plasmids from above. There must always be a pStG plasmid as well as either pFRYC or pFRY. However, additional controls can be conducted using only one of the three plasmids. | To use the kit properly, each culture must have two of the three plasmids from above. There must always be a pStG plasmid as well as either pFRYC or pFRY. However, additional controls can be conducted using only one of the three plasmids. | ||
| Line 147: | Line 150: | ||
*Actual synthetase/tRNA pairs can also sometimes have '''low efficiency'''. As these are artificially selected pairs, sometimes they do not function as well as natural synthetase/tRNA pairs. In such a case, the amount of sfGFP translated in Figure 3D might be significantly lower than expected. '''low efficiency''' is also not ideal as it may limit levels of protein expression. | *Actual synthetase/tRNA pairs can also sometimes have '''low efficiency'''. As these are artificially selected pairs, sometimes they do not function as well as natural synthetase/tRNA pairs. In such a case, the amount of sfGFP translated in Figure 3D might be significantly lower than expected. '''low efficiency''' is also not ideal as it may limit levels of protein expression. | ||
| - | + | <h2>Experimental Preparation</h2> | |
| Line 184: | Line 187: | ||
| - | + | <h1>Results and Data</h1> | |
| - | + | <h2>Fidelity of Incorporation</h2> | |
[[File:UT_Austin_2014_Kit_Normalized_GFP_to_RFP_graph.png|thumb|600px|Figure 3. Graph showing the level of GFP fluorescence relative to RFP fluorescence for each condition. Each pStG plasmid is referred to based on the tRNA synthetase/tRNA pair present in the specific plasmid. Each of these plasmids was then paired with either pFRY or pFRYC and grown in the presence or absence of a specific ncAA. For example, the "3-AminoY-FRYC" and the "3-AminoY-FRY" samples both contain the 3-AminoY synthetase/tRNA pair and both samples were grown in the absence or presence of the ncAA "3-AminoY". Data are presented as the average of three independent cultures. Error bars denote standard deviation.]] | [[File:UT_Austin_2014_Kit_Normalized_GFP_to_RFP_graph.png|thumb|600px|Figure 3. Graph showing the level of GFP fluorescence relative to RFP fluorescence for each condition. Each pStG plasmid is referred to based on the tRNA synthetase/tRNA pair present in the specific plasmid. Each of these plasmids was then paired with either pFRY or pFRYC and grown in the presence or absence of a specific ncAA. For example, the "3-AminoY-FRYC" and the "3-AminoY-FRY" samples both contain the 3-AminoY synthetase/tRNA pair and both samples were grown in the absence or presence of the ncAA "3-AminoY". Data are presented as the average of three independent cultures. Error bars denote standard deviation.]] | ||
| Line 196: | Line 199: | ||
To determine the change in GFP fluorescence when the ncAA was present, we first had to calculate how much GFP was expressed relative to the RFP, which would give an upper estimate of how much GFP could theoretically be expressed. We first divided both the GFP and RFP levels by the OD 600 of the culture in order to get the per cell fluorescence levels. We then normalized the GFP fluorescence for one culture to its RFP fluorescence so that we could compare the GFP fluorescence levels between cultures. The normalized GFP values were then compared between cultures grown in the presence of ncAA and cultures grown in the absence of ncAA, which would indicate how the level of GFP fluorescence changes when the ncAA is present. When these values were graphed (Figure 3), some synthetase/tRNA pairs such as 4-azidophenylalanine, 3-nitrotyrosine, 3-iodotyrosine, and ortho-nitrobenzyltyrosine resulted in higher GFP fluorescence in the presence of ncAA than in the absence of ncAA, which suggests that those synthetases only incorporated an amino acid if their specific amino acid was present, meaning that they have a high fidelity. However, the other synthetase/tRNA pairs (3-aminotyrosine, L-DOPA, and cyanophenylalanine) did not show a significant increase in GFP fluorescence normalized to RFP fluorescence when the ncAA was present, indicating that they would incorporate other amino acids at the amber stop codon when their specific amino acid was not present (and perhaps even when it was), and thus have a low fidelity. | To determine the change in GFP fluorescence when the ncAA was present, we first had to calculate how much GFP was expressed relative to the RFP, which would give an upper estimate of how much GFP could theoretically be expressed. We first divided both the GFP and RFP levels by the OD 600 of the culture in order to get the per cell fluorescence levels. We then normalized the GFP fluorescence for one culture to its RFP fluorescence so that we could compare the GFP fluorescence levels between cultures. The normalized GFP values were then compared between cultures grown in the presence of ncAA and cultures grown in the absence of ncAA, which would indicate how the level of GFP fluorescence changes when the ncAA is present. When these values were graphed (Figure 3), some synthetase/tRNA pairs such as 4-azidophenylalanine, 3-nitrotyrosine, 3-iodotyrosine, and ortho-nitrobenzyltyrosine resulted in higher GFP fluorescence in the presence of ncAA than in the absence of ncAA, which suggests that those synthetases only incorporated an amino acid if their specific amino acid was present, meaning that they have a high fidelity. However, the other synthetase/tRNA pairs (3-aminotyrosine, L-DOPA, and cyanophenylalanine) did not show a significant increase in GFP fluorescence normalized to RFP fluorescence when the ncAA was present, indicating that they would incorporate other amino acids at the amber stop codon when their specific amino acid was not present (and perhaps even when it was), and thus have a low fidelity. | ||
| - | + | <h2>Synthetase Efficiency</h2> | |
Another measure of quality for these ncAA synthetase/tRNA pairs is how efficiently they incorporate their ncAA. An inefficient synthetase/tRNA pair will only incorporate their ncAA a fraction of the time, even when their ncAA is present. Our system can be used to measure this level of efficiency, though it does not indicate why a particular synthetase is efficient or inefficient. By comparing the level of GFP fluorescence to the normalized level of RFP fluorescence when the amino acid is present, we can see how efficient the synthetase is. In essence, if the normalized fluorescence of GFP relative to RFP is close to 100% (if the GFP is expressed roughly 100% of the time that RFP is expressed), then the synthetase is very efficient. On the other side, if say the normalized fluorescence of GFP relative to RFP is closer to 10%, then the synthetase would not be very efficient, because even when the ncAA was there, it only incorporated it at the amber stop codon (UAG) about 10% of the time. | Another measure of quality for these ncAA synthetase/tRNA pairs is how efficiently they incorporate their ncAA. An inefficient synthetase/tRNA pair will only incorporate their ncAA a fraction of the time, even when their ncAA is present. Our system can be used to measure this level of efficiency, though it does not indicate why a particular synthetase is efficient or inefficient. By comparing the level of GFP fluorescence to the normalized level of RFP fluorescence when the amino acid is present, we can see how efficient the synthetase is. In essence, if the normalized fluorescence of GFP relative to RFP is close to 100% (if the GFP is expressed roughly 100% of the time that RFP is expressed), then the synthetase is very efficient. On the other side, if say the normalized fluorescence of GFP relative to RFP is closer to 10%, then the synthetase would not be very efficient, because even when the ncAA was there, it only incorporated it at the amber stop codon (UAG) about 10% of the time. | ||
| Line 203: | Line 206: | ||
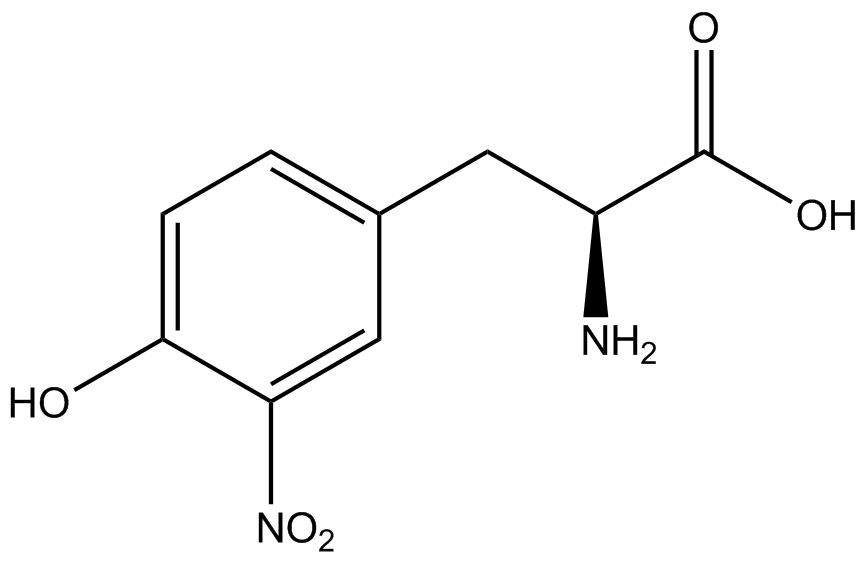
For our results (Figure 3), two synthesase/tRNA pairs stood out as relatively inefficient: 3-nitrotyrosine and ortho-nitrobenzyltyrosine. Both of these synthetase/tRNA pairs showed a significantly smaller normalized GFP to RFP fluorescence when the amino acid was present with the pFRY construct. While they both show a significant increase in normalized GFP to RFP fluorescence when the amino acid was present compared to when it was absent, which indicates a high fidelity, the actual GFP fluorescence relative to the RFP fluorescence was only around 50% for 3-nitrotyrosine and 20% for ortho-nitrobenzyltyrosine. These results suggest that for whatever reason, these synthetase/tRNA pairs do not always incorporate their ncAA at the amber stop codon. | For our results (Figure 3), two synthesase/tRNA pairs stood out as relatively inefficient: 3-nitrotyrosine and ortho-nitrobenzyltyrosine. Both of these synthetase/tRNA pairs showed a significantly smaller normalized GFP to RFP fluorescence when the amino acid was present with the pFRY construct. While they both show a significant increase in normalized GFP to RFP fluorescence when the amino acid was present compared to when it was absent, which indicates a high fidelity, the actual GFP fluorescence relative to the RFP fluorescence was only around 50% for 3-nitrotyrosine and 20% for ortho-nitrobenzyltyrosine. These results suggest that for whatever reason, these synthetase/tRNA pairs do not always incorporate their ncAA at the amber stop codon. | ||
| - | + | <h2>Incorporation Value</h2> | |
[[File:UT_Austin_2014_Kit_Incorporation_Value_Graph.png|600px|thumb|Figure 4.<br> | [[File:UT_Austin_2014_Kit_Incorporation_Value_Graph.png|600px|thumb|Figure 4.<br> | ||
<i>May need to consider a different naming convention or the cultures. Possibly AY-C and AY-E for Control and Experimental?? </i> The fluorescence and OD600 readings of each culture were used to calculate a value for incorporation efficiency of each synthetase. <i>Need to add an explanation of how these values were calculated</i>]] | <i>May need to consider a different naming convention or the cultures. Possibly AY-C and AY-E for Control and Experimental?? </i> The fluorescence and OD600 readings of each culture were used to calculate a value for incorporation efficiency of each synthetase. <i>Need to add an explanation of how these values were calculated</i>]] | ||
| Line 217: | Line 220: | ||
| - | + | <h1>Discussion</h1> | |
'''What's your most important result?''' DISCUSS THAT FINDING FIRST. | '''What's your most important result?''' DISCUSS THAT FINDING FIRST. | ||
| Line 239: | Line 242: | ||
'''ADD MASS SPEC COMMENTARY''' | '''ADD MASS SPEC COMMENTARY''' | ||
| - | + | <h1>Conclusion</h1> | |
This year the UT Austin iGEM Team has chosen to take part in the [https://2014.igem.org/Tracks/Measurement Measurement Track] of the iGEM competition by contributing to ncAA methods of analysis and fulfilling [https://2014.igem.org/Team:Austin_Texas/medal_requirements medal criteria]. Our team has successfully designed a unique testing kit for ncAA incorporation <i>in vivo</i> using common reporter parts to characterize very uncommon synthetase/tRNA machinery. In addition, we have used our kit to analyze and characterize seven different ncAA synthetase/tRNA pairs as well as tyrosine RS. This method of testing was designed to be undergraduate administrable, transportable, and more affordable than other methods of characterization and analysis. | This year the UT Austin iGEM Team has chosen to take part in the [https://2014.igem.org/Tracks/Measurement Measurement Track] of the iGEM competition by contributing to ncAA methods of analysis and fulfilling [https://2014.igem.org/Team:Austin_Texas/medal_requirements medal criteria]. Our team has successfully designed a unique testing kit for ncAA incorporation <i>in vivo</i> using common reporter parts to characterize very uncommon synthetase/tRNA machinery. In addition, we have used our kit to analyze and characterize seven different ncAA synthetase/tRNA pairs as well as tyrosine RS. This method of testing was designed to be undergraduate administrable, transportable, and more affordable than other methods of characterization and analysis. | ||
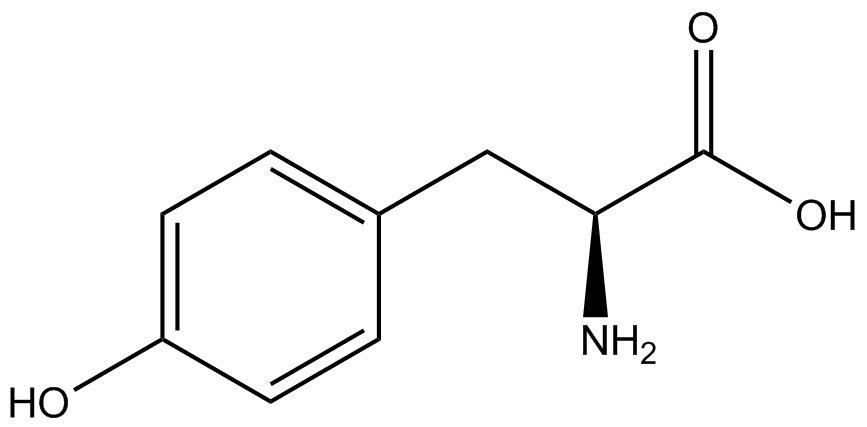
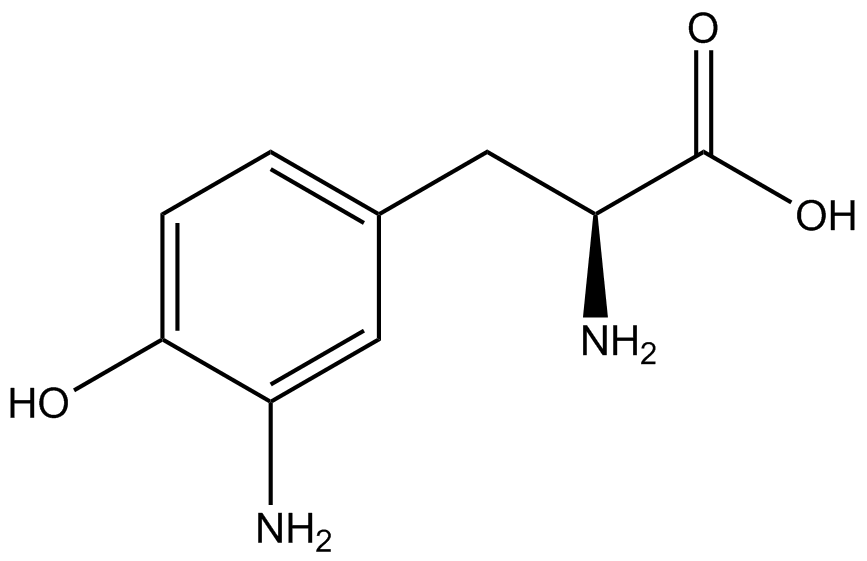
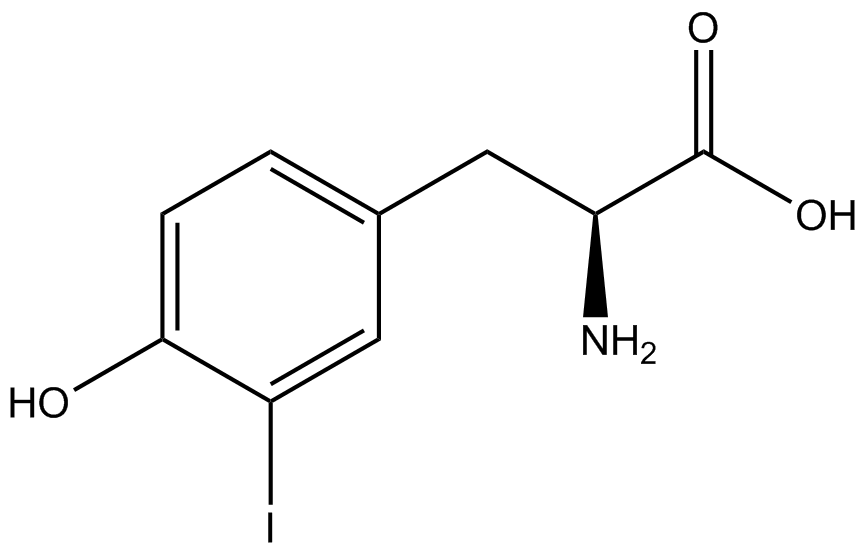
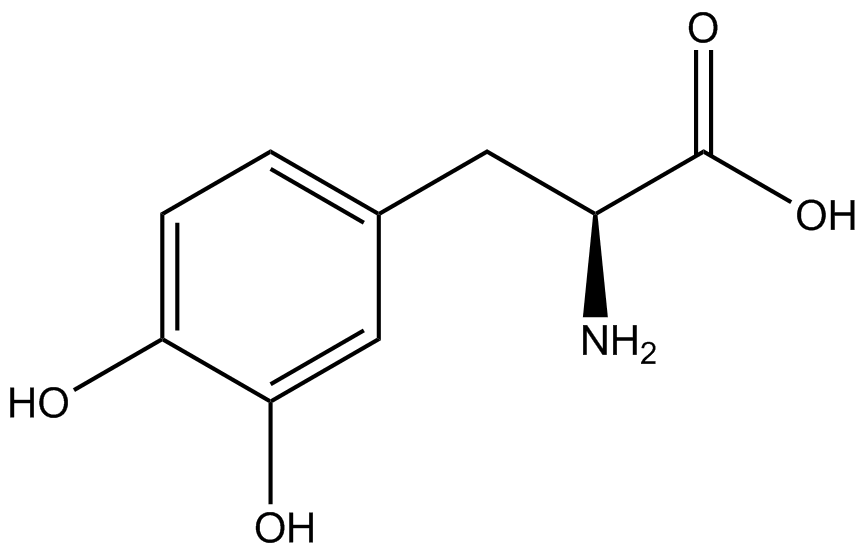
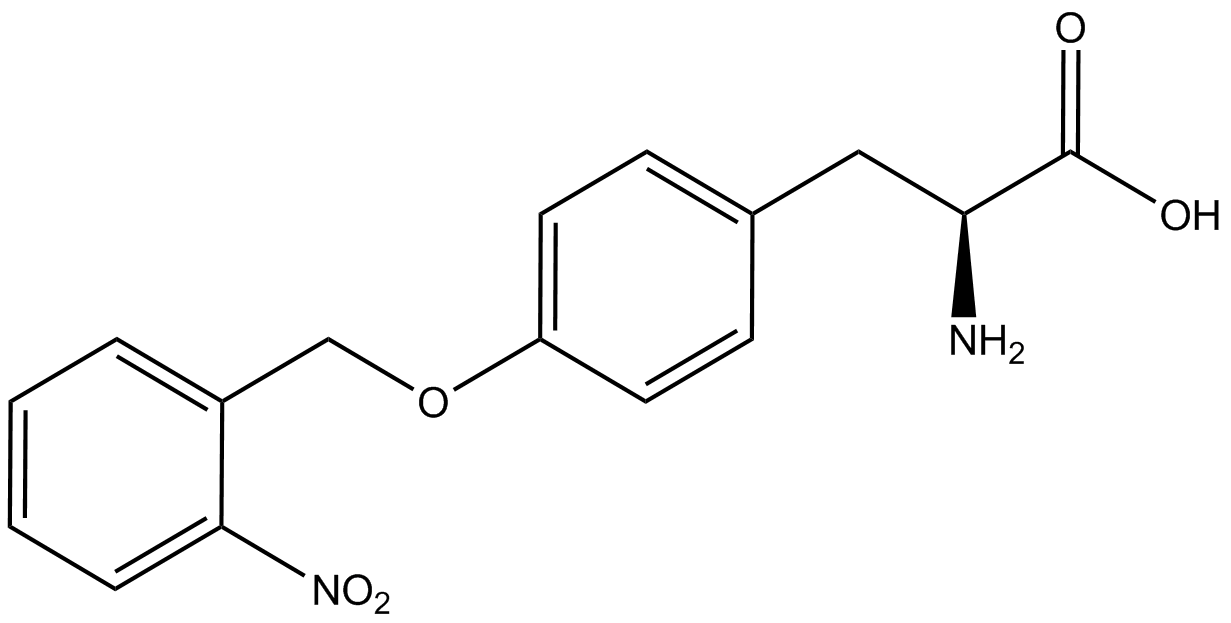
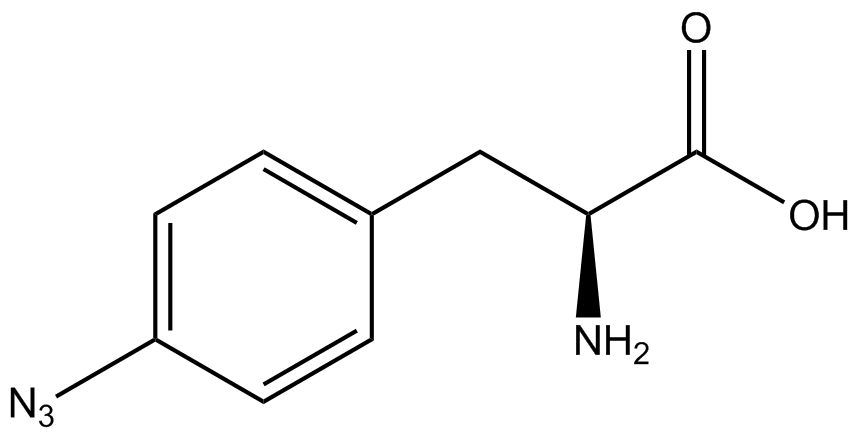
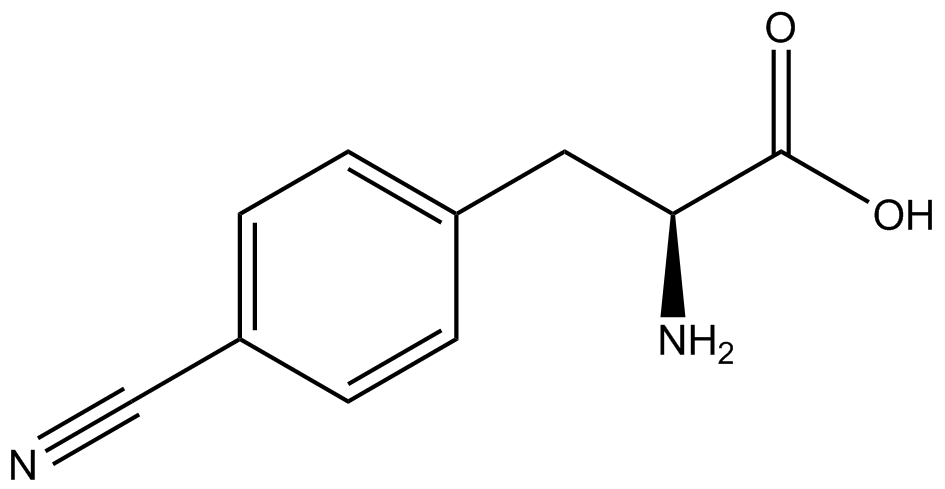
| - | + | <h1>ncAA Table</h1> | |
| Line 313: | Line 316: | ||
|} | |} | ||
| - | + | <h1>ncAA Synthetase Table</h1> | |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 398: | Line 401: | ||
|} | |} | ||
| - | + | <h1>References</h1> | |
* Alfonta, L., Zhang, Z., Uryu, S., Loo, J. A., Schultz, P. G. (2003) Site-specific incorporation of a redox-active amino acid into proteins. ''J. Am. Chem. Soc.'' '''125''':14662-14663. | * Alfonta, L., Zhang, Z., Uryu, S., Loo, J. A., Schultz, P. G. (2003) Site-specific incorporation of a redox-active amino acid into proteins. ''J. Am. Chem. Soc.'' '''125''':14662-14663. | ||
Revision as of 02:49, 17 October 2014
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"