Team:RHIT/Project/HumanPractices
From 2014.igem.org
Overview
Synthetic biology is a rapidly emerging field in the world and has been getting a lot of attention in the media recently. Even with all of the press that different forms of synthetic biology have been getting, there is actually little that is known about this newly emerging field. For this reason, the Rose-Hulman Institute of Technology iGEM team created Victor the Vector to aid in teaching the youth of the world about synthetic biology.
Victor the Vector is a hands-on interactive device that allows students to build basic synthetic biological systems and watch instructional videos about the systems that were built. This way students can experience building synthetic biological systems without the need of a wet lab or any wet lab experience. This is done by modeling the shape and design of Victor the Vector as a plasmid. The figure below shows the design of Victor the Vector.
As shown in the picture of Victor the Vector above, there are twelve slots in which different parts of a synthetic biological system can be placed. This way all the user has to do is take the provided parts and plug them into Victor the Vector in the correct order to produce a viable synthetic biology system. This is meant to mimic how synthetic biologists take different genetic parts and put them together in a novel way on a plasmid. If the student has plugged the correct pieces into Victor the Vector in the correct order then it will initiate an instructional video which the student can watch to learn more about the system that he or she has built. This way students get a hands-on approach with building synthetic biological systems and a basic understanding of some of the components and the order in which parts need to be organized to create a synthetic biological system. Our hope is that by giving the students a device to play and interact with, they will gain a better appreciation and understanding of this nanoscale science.
-
Victor the Vector is an electromechanical device that allows students to practice building synthetic biological systems that are programmed into the device. This way if the user builds the system correctly, the device will send a signal to the student's computer which will activate an instructional video. This is achieved by giving each plug in part a unique voltage so that every system that is built has a unique series of voltages at the output of the device. See the figure below for a schematic of the wiring in the device.
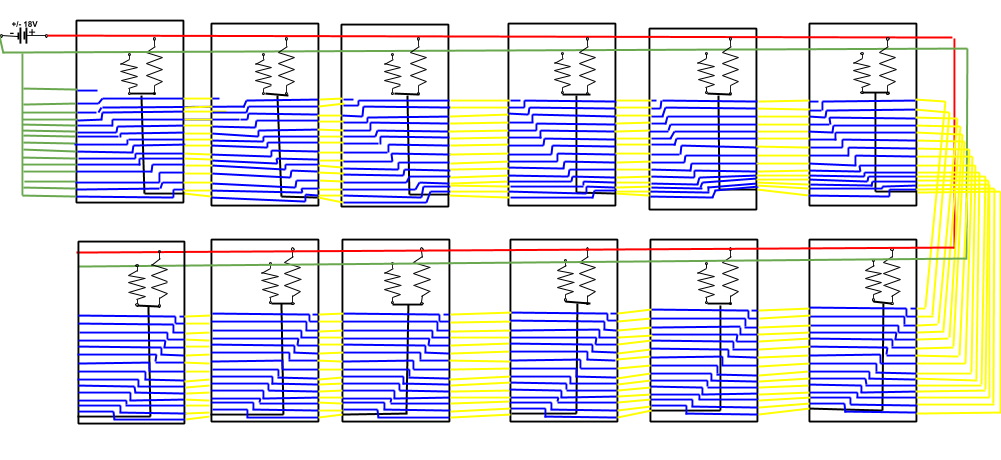
Two onboard NI-DAQs convert the signal from analog to digital. The digital signal is then sent from the NI-DAQs to the user's computer via usb. The digital data is then imported into Matlab where it is processed and used to initiate the instructional video that is associated with the system that was built. This way the user gets instant feedback on whether or not they have built the system properly. Currently there are only five synthetic biological systems pre-programmed into the device. However, the device has been designed to allow for addition of more pre-programmed synthetic biological systems for future models.
-
Part Description 
General Promoter (2)
General promoters are regions of DNA that provide specific DNA sequences to which RNA polymerase and transcription factors can bind, thus initiating transcription.

Lac Promoter
The lactate (lac) promoter is inducible, which means that it is turned "on" and allows for RNA polymerase binding and transcription initiation when lactate is present in the cell.

FUS1 Promoter
The FUS1 promoter is a pheromone-inducible yeast promoter. A gene of interest can be expressed when the FUS1 promoter is induced by the appropriate yeast mating pheromone.

Operator
An operator is a segment of DNA to which a protein called a transcription factor binds. Operators are key in regulating gene expressoin; repressor and inducer proteins can bind and either inhibit or facilitate transcription.

LexA Regulatory Element
This regulatory element is the promoter to which the LexA binding domain and VP64 activator domain bind. This facilitates further production of the blue heteroprotein.

LexA Biding Domain
This binding domain forms a fusion with the VP64 activator and binds to the LexA regulatory element, facilitating further production of the blue heteroprotein.

Alpha Mating Factor
Yeast cells can be one of two different mating types, a or α, which determines the sexual behavior of the cell. This mating pheromone α-factor is produced by α strain yeast, and is recognized by a strain cells, which respond to α-factor by growing a projection called a shmoo towards the source of α-factor.

HIS3
HIS3 is a gene found in yeast; it is analogous to HisB in E. coli. This gene encodes for a protein that catalyzes the sixth step in histidine biosynthesis. This gene is sometimes used for selection purposes.

Terminator (2)
Terminators mark the end of a gene in genomic DNA. They are a section of DNA sequence that provides signals to the newly synthesized mRNA that trigger processes that release mRNA from the transcriptional complex, thus terminating translation.

RBS (2)
A ribosomal binding site is a sequence of mRNA that is bound by the ribosome when initiating protein translation. The ribosome searches for this site and binds to it via base pairing. The ribosome can then begin translation.

VP64
Viral Protein 64 is a transcriptional activator composed of four copies of a Herpes Simplex Viral Protein connected by glysine-serine linkers. When fused with a protein domain that can bind near the promoter of a gene, such as LexA, VP64 acts as a strong transcriptional activator. This serves to continue the expression of the BFP genes.

INPNC
This gene codes for the ice nucleation protein found in Xanthomonas campestris. Its function is to provide a surface for ice crystal formation, but here it is utilized for its surface display properties to display mating factor α on the surface of
coli. 
LDH
This gene codes for lactate dehydrogenase, which catalyzes the interconversion of pyruvate and lactate. The lactate produced can then be used to induce a lactate promoter, such as the one in the E. coli construct.

BFP
This blue fluorescent protein gene is derived from the jellyfish. This gene and genes like it, including green fluorescent protein (GFP), are frequently used to report expression; in the latch circuit presented here, it is used to report expression of the mating factor α gene.

GSL
The glysine-serine linker is a short peptide sequence that occurs between protein domains to hold the domains together. Flexible residues, such as glysine and serine, are used to allow adjacent protein domains to move freely. In the case of the latch circuit, the GSLs connect the BFP domains.

NLS
A nuclear localization signal is an amino acid sequence that serves to “tag” a protein for import into the nucleus.

LacI
This gene codes for the lac repressor. When lactose is not present, the lac repressor binds to the operator region of the lac operon, which prevents transcription of the mRNA coding for the lac proteins needed to digest lactose.

LAC ZYA
These are the structural genes of the lac operon. They encode the enzymes necessary to break down lactose. These enzymes are only produced when lactose is present (see LAC I above).

trpR
This gene encodes the repressor protein for the trp operon, and is continually expressed at a low level. When tryptophan is present it binds to the repressor, which then binds to the operator and prevents transcription, thus inhibiting tryptophan production.

trpL
Coding for the trp operon leader transcript is included in trpL. Embedded in trpL is the transcript for secondary structures that control attenuation, which is a mechanism of negative feedback.

Attenuator Sequence
The attenuator is a provisional stop signal located in the DNA segment that corresponds to the leader sequence of mRNA. The ribosome becomes stalled in this attenuator region, and transcription either stops or is allowed to go ahead to the structural gene part of the mRNA.

trp EDCBA
These are the structural genes of the trp operon. They code for tryptophan synthetase, which is responsible for catalyzing steps in tryptophan synthesis.

Filler Blocks
These pieces have no biological significance. Their purpose is to fill the empty spaces that follow smaller synthetic systems, in order to complete the electric circuit.
-
To view our Ethics pamphlet click here!
-
To view our Business Plan click here!
 "
"


